Faculty
What Lies Within
The new, ultra-resolution nanoscope gives biological researchers the power to see deep within whole cells.

Until recently, the challenge with optical microscopy has been to obtain complete, high resolution, images of cells and tissues because resolution rapidly deteriorates due to the thickness of the structures. With a microscope that now allows three-dimensional imaging of whole cells, it is now possible to glimpse what lies within them. Whitney Duim ’05, a chemistry alumna and visiting assistant professor in the Harvey Mudd College Department of Chemistry, was involved in developing the new microscope as a post-doctoral scholar at Yale University. Based on this work, Duim and her colleagues published the article, “Ultra-High Resolution 3-D Imaging of Whole Cells,” in the journal Cell.
The new microscope—whole-cell 4Pi singlemolecule switching nanoscopy (W-4PiSMSN)— represents the culmination of more than a decade’s research on high-resolution fluorescence imaging techniques. And, it establishes 3-D biological imaging with molecular specificity and resolution in the 10-nanometer range as a general imaging technique.
“Development of this new microscope makes it possible to get to 10-20 nanometers resolution in three dimensions and see the entire structure of cells,” Duim says. “A lot of biological processes happen at the nanometer scale. Being able to see that in three dimensions, high-resolution is a big deal! There are going to be a lot of researchers who are going to want one of these microscopes.”
Duim developed the preparation of samples and took the first images of three different biological structures—microtubule filaments, clathrin-coated vesicles and the Golgi apparatus in mammalian cells—on the nanoscope. After taking the photos, the team used a computer algorithm to process the data and was able to see the structures in three dimensions. This served as the first proof-ofprinciple for the team.
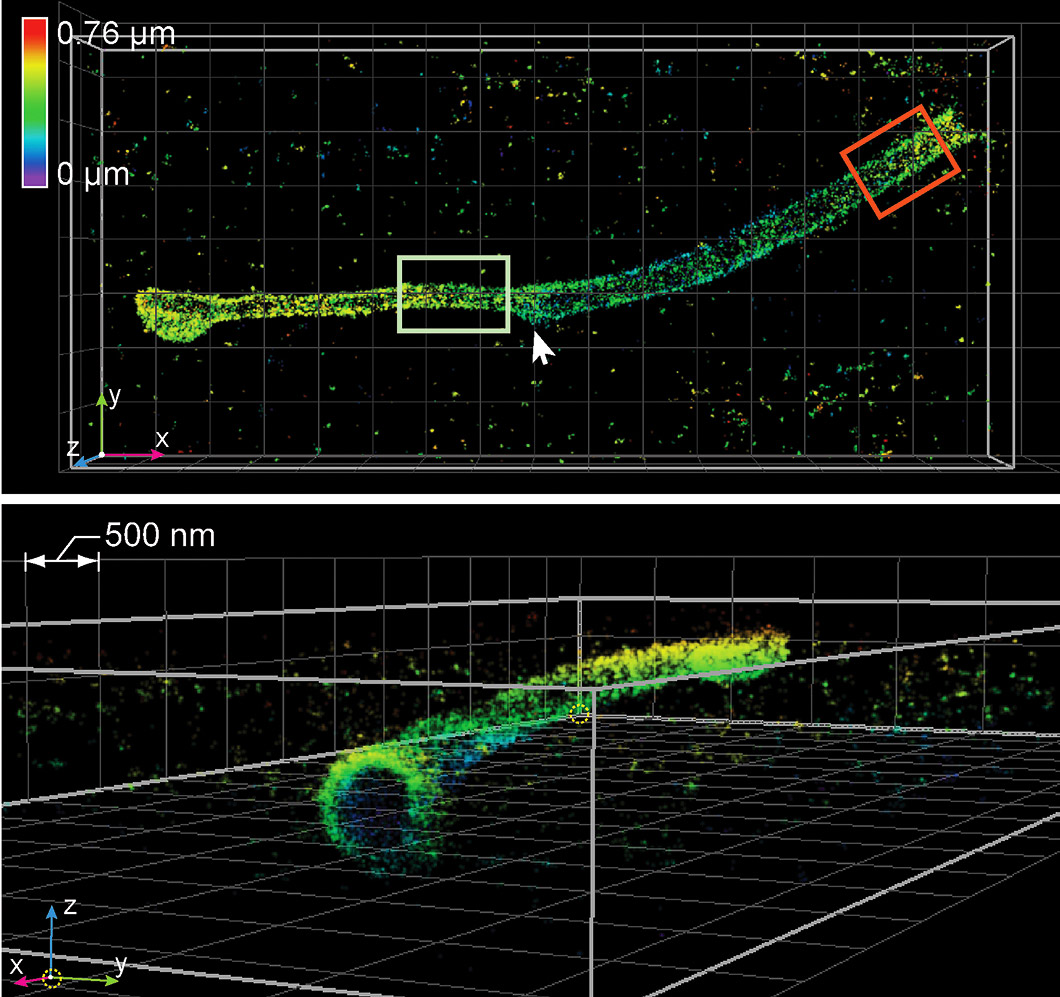
“It’s a very complicated microscope that took a long time to build,” Duim says. “So getting those first images [was] very exciting.”
Two major advances made it possible to take photos at all depths in a cell. Light-collecting objective lenses were placed both underneath and on top of the sample. Then, the team applied adaptive optics, a technology used commonly in astronomy, to correct for distortion of light as it passed through the cells.
“We hope the microscope will unlock other advances in biology, basically being able to capture more interaction in the cells, more structures in the cell and contribute to our understanding not only of what is going on in the cell but [potentially assist in] therapeutic and medical applications,” Duim says. Unlike other powerful microscopes, this one also has the potential to provide high-resolution imaging of living cells.
“There’s a tremendous amount of very important information you can get from fixed cells. But with this technology, we hope to be able to watch movement in live cells in real time in high resolution,” she says.
Duim is sharing her knowledge of this important instrument with Harvey Mudd students. During the past school year and summer, Duim and her student researchers built a custom, two-dimensional, highresolution microscope. They studied the aggregation of the protein involved in Huntington’s disease, a progressive, neurodegenerative disease that is fatal and incurable.
“Our microscope is not as fancy as the threedimensional instrument, but we can do [highresolution] two-dimensional imaging quite well,” Duim says. Their summer research was funded by the HMC chemistry department, a grant from The Claremont Colleges’ Collaborative HHMI Undergraduate Science Education Award to Duim and Professor Rachel Levy of the HMC Department of Mathematics, and the Norman F. Sprague III, M.D. Experiential Learning Fund established by the Jean Perkins Foundation.
Carla Becker ’18, Rebecca Harman ’17 and Ali Khan ’19 worked with Duim over the summer creating the protein from DNA, preparing the aggregates from that protein and then recording and analyzing the images. Becker and Harman are continuing work with Duim this school year. They are studying how the aggregates grow, their structure and how sensitive they are to the environment in which they form.
“The hope is eventually to be able to identify therapeutic [targets], ways to inhibit aggregation or cause formation of aggregate species that are not as toxic to the cell,” Duim says.